Genetic Testing
Is there any genetic testing available for ASD?

No genetic testing is currently available for the diagnosis of autism. Autism is diagnosed by behavioral observations of core deficits by trained professionals according to the guidelines established by the Diagnostic and Statistical Manual of Mental Disorders (DSM IV –TR).
However, some genetic tests already exist to guide ASD medical management. At present, these tests can only identify a genetic risk factor in 15-20% of ASD individuals that undergo testing. New techniques such as next generation sequencing (NGS) and chromosomal microarray analysis (CMA) show promising results for identifying genetic causes in a larger percentage of ASD individuals because of their ability to analyze DNA at higher resolution. NGS is typically used to identify changes in both single nucleotides (also known as single nucleotide variants, or SNVs) and small insertions or deletions of DNA. CMA is generally used to identify large chromosomal gains or losses, also known as copy number variants (CNVs), but one type of CMA method called an SNP array can also detect single nucleotide changes. Notably, NGS technologies can also be used to identify CNVs, as was shown in a recent study of autistic individuals1.
In 2008, the American College of Medical Genetics developed practice guidelines to aid clinicians with genetic testing related to ASD2.
What is next generation sequencing (NGS)?
Next generation sequencing (NGS) is a term used to describe a collection of high-throughput sequencing technologies that enable clinicians to screen larger amounts of genetic material at a lower cost than traditional sequencing technologies. These technologies have greatly expanded the number of potentially pathogenic variants in individuals affected by a wide range of diseases. Three principal NGS approaches can potentially be used to identify disease-associated variants in the genomes of ASD individuals: targeted gene panels, whole exome sequencing, and whole genome sequencing. Exome sequencing has already been used to identify variants in ASD individuals1, 3-7.
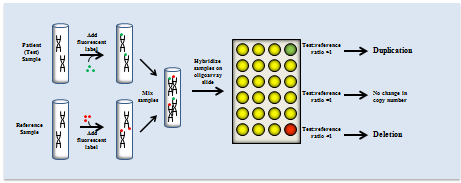
What is chromosomal microarray (CMA) screening?
Given the evidence that copy number variants (CNVs) are important genetic risk factors for ASD, increasingly more ASD individuals are undergoing genetic screening for potentially pathogenic CNVs across their genome, a procedure known as chromosomal microarray (CMA) screening. The term CMA is frequently used to include all types of array-based genomic copy number analyses, such as array-comparative genomic hybridization (aCGH) and single nucleotide polymorphism (SNP) arrays. CMA screening has been proposed to replace traditional cytogenetic testing as a first-tier diagnostic test in individuals with ASD and other developmental disabilities10.
| References: |
|