What is a copy number variant, and why are they important risk factors for ASD?
For decades, scientists have known that microscopically visible chromosomal rearrangements can result in a wide range of developmental disorders. However, technological and computational advances in the past decade have enabled the development of assays capable of identifying even smaller structural changes in chromosomes that could not have been detected by traditional cytogenetic analysis.
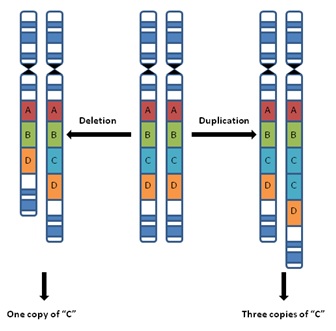
Among the most heavily scrutinized of these submicroscopic structural variants are copy number variants, or CNVs. CNVs are submicroscopic chromosomal deletions and/or duplications that are typically defined as DNA segments 1000 basepairs or larger in size that are present in a different number of copies when compared to a reference (or standard) genome1. For example, as shown in the middle of the figure below, the pair of normal chromosomes each have sections A-B-C-D. However, the loss of section C from one of the chromosomes results in an abnormal chromosome with only sections A-B-D (left pair in the figure below); an individual with this deletion therefore now has only one copy of section C in their chromosomes.
On the other hand, the gain of an extra copy of section C on one of the chromosomes results in an abnormal chromosome with sections A-B-C-C-D (right pair in the figure below); an individual with this duplication now has three copies of section C in their chromosomes. In other words, both of these individuals (left and right in the figure below) have CNVs involving section C – one has lost a copy, the other has gained a copy, but both have a varied number of copies of C when compared to the original, reference set of chromosomes.
| References: |
|

